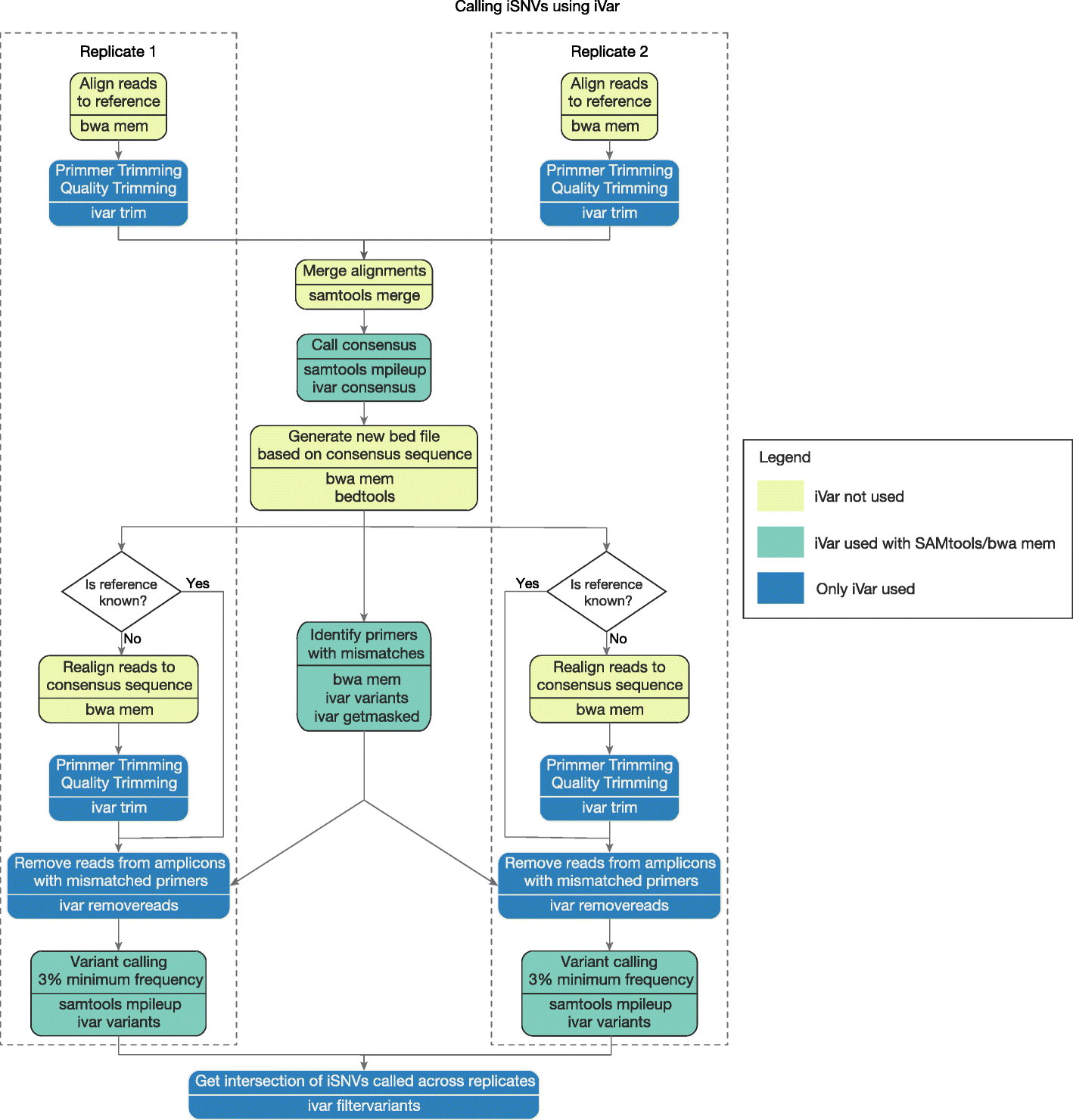
Cookbook
Two pipelines to call iSNVs from known and unknown reference sequences have been written in snakemake.
The two pipelines are distributed along with iVar and are present in thepipeline/ and pipeline_field/ fodlers respectively.
For both pipelines, there are four parameters that will have to be set in beginning of the Snakefile.
in_dir = "<input fastq files>"
out_dir = "<output directory>"
bed = "<bed-file-with-primer-positions>"
ref="<path to reference fasta>"
 1.10.0
1.10.0